Linear Discriminant Analysis¶
We introduce Fisher’s Linear Discriminant Function.
Objective¶
We have two groups of data in \(\mathbb{R} ^p\)
\(\left\{ \boldsymbol{x} _{1i} \right\}, i = 1, 2, \ldots, n_1\)
\(\left\{ \boldsymbol{x} _{2i} \right\}, i = 1, 2, \ldots, n_2\)
We want to project them to a one dimensional line in \(\mathbb{R}\), such that the projection by this line yields good separation of the two classes.
Let the projection be \(\boldsymbol{a} ^{\top} \boldsymbol{x}\) and let the projected data be
\(\left\{ y _{1i} = \boldsymbol{a} ^{\top} \boldsymbol{x} _{1i} \right\}\)
\(\left\{ y_{2i} = \boldsymbol{a} ^{\top} \boldsymbol{x} _{2i} \right\}\)
The goodness of separation is measured by th ratio of between-class difference and within-class variance.
where \(s_y^2\) is the pooled variance, if we assume equal covariance structure in the two groups.
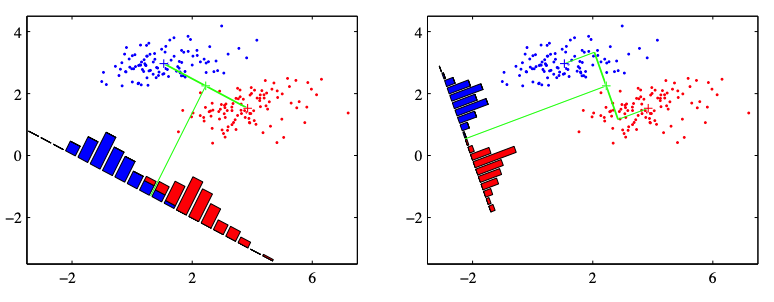
Fig. 83 Illustration of projection of data to a linear direction [C. Bishop, 2006]¶
Assumptions¶
equal covariance matrix \(\boldsymbol{\Sigma} _1 = \boldsymbol{\Sigma} _2 = \boldsymbol{\Sigma}\)
full rank \(\operatorname{rank}\left( \boldsymbol{\Sigma} \right) = p\)
without normality assumption that the population are from multivariate normal.
Learning¶
Note that the objective function
has the form of generalized Rayleigh quotient. Hence the solution is given by
with maximum
The maximum \(D^2\) can be viewed as the square of the Mahalanobis distance between the population means of the original data.
Prediction¶
For a new data point \(\boldsymbol{x} _0\), we compute \(y_0 = \boldsymbol{a} ^{* \top } \boldsymbol{x} _0\), and see whether \(y_0\) is closer to \(\bar{y}_1\) or \(\bar{y}_2\). Below are two equivalent classification rules.
Define the midpoint of the transformed means \(m = \frac{1}{2}(\bar{y}_1 + \bar{y}_2)\) as the partition point, suppose \(\bar{y}_1 \ge \bar{y}_2\), then
\[ R_1: \quad y_0 \ge \frac{1}{2}(\bar{y}_1 + \bar{y}_2) \]Assign it to closer class, in terms of absolute distance
\[j^* = \min_{j=1, 2} \left\vert y_0 - \bar{y}_j \right\vert\]Assign it to closer class, in terms of squared distance
\[ R_1: \quad (y_0 - \bar{y}_1)^2 \le (y_0 - \bar{y}_2)^2 \]or using \(\boldsymbol{x}\)
\[ R_1: \quad \left\vert \boldsymbol{a} ^{* \top} (\boldsymbol{x} _0 - \bar{\boldsymbol{x} }_1) \right\vert^2 \le \left\vert \boldsymbol{a} ^{* \top} (\boldsymbol{x} _0 - \bar{\boldsymbol{x} }_2) \right\vert^2 \]
Multi-classes¶
We can extend two classes to multi-classes, and extend projection to one line to projection to some dimensions, i.e. as a dimensionality reduction method.
The assumptions remain the same
equal covariance matrix \(\boldsymbol{\Sigma} _1 = \ldots = \boldsymbol{\Sigma} _g = \boldsymbol{\Sigma}\)
full rank \(\operatorname{rank}\left( \boldsymbol{\Sigma} \right) = p\)
without normality assumption that the population are from multivariate normal.
Learning¶
We first consider projection to one line case. The objective is to maximize
where
between-group variance matrix (different from \(\boldsymbol{B}\) in MANOVA)
\[\boldsymbol{B}=\sum_{i=1}^{g}\left(\bar{\boldsymbol{x}}_{i}-\bar{\boldsymbol{x}}\right)\left(\bar{\boldsymbol{x}}_{i}-\bar{\boldsymbol{x}}\right)^{\top}\]mean-of-means, aka overall average, in contrast to grand average
\[\bar{\boldsymbol{x}} = \frac{1}{g} \sum_{i=1}^g \bar{\boldsymbol{x}}_i\]within-group variance matrix (same \(\boldsymbol{W}\) in MANOVA)
\[\boldsymbol{W}=\sum_{i=1}^{g}\left(n_{i}-1\right) \boldsymbol{S}_{i}=\sum_{i=1}^{g} \sum_{j=1}^{n_{i}}\left(\boldsymbol{x}_{i j}-\bar{\boldsymbol{x}}_{i}\right)\left(\boldsymbol{x}_{i j}-\bar{\boldsymbol{x}}_{i}\right)^{\top}\]
This is again a generalized Rayleigh quotient. The optimizer \(\boldsymbol{a} ^*\) and maximum is given by the largest eigen pair of matrix \(\boldsymbol{W} ^{-1} \boldsymbol{B}\).
Recall that \(\boldsymbol{S} _{\text{pool} } = \frac{1}{n-g} \boldsymbol{W}\). The eigenvector \(\boldsymbol{a} ^*\) is usually chosen such that
After obtain one projection direction \(\boldsymbol{a} _1 ^*\), we can continue for next, i.e. second largest eigenvector, and so on. There eigenvectors \(\boldsymbol{a} _j ^*\) are called discriminant coordinates, or canonical variates, coming from an alternative derivation via CCA on predictor variable matrix and response variable matrix.
Note that there are at most \(r\) non-zero eigenvalues of \(\boldsymbol{W} ^{-1} \boldsymbol{B}\) where
Therefore the above method can produce up to \(r\) linear discriminants.
By the maximum procedure and proper scaling, we have
Interpretation¶
They give consecutive directions that maximized the normalized variance between classes, they are not discriminant function themselves. Analogous to the principal components in PCA, hopefully the first few sample discriminants show better separation of the groups, and can be used to classify new observations.
Prediction¶
Recall that for \(g=2\) case, the classification rule using squared distance is
In case of \(r\) discriminant coordinates \(\boldsymbol{a} _1, \ldots, \boldsymbol{a} _r\), we allocate \(\boldsymbol{x} _0\) to class \(k\) if
Equivalently,
where \(\boldsymbol{A} \in \mathbb{R} ^{p \times r}\) and consists \(r\) scaled eigenvectors such that \(\boldsymbol{A} ^{\top} \boldsymbol{S} _{\text{pool} }\boldsymbol{A} = \boldsymbol{I} _r\).
R.t. Discriminant for Gaussian Data¶
In the \(g=2\) case above, we showed that Fisher LDA is equivalent to the linear discriminant rule for Gaussian that minimizes ECM under some assumptions. Now we show they are equivalent in \(g \ge 2\) case, under those assumptions.
Recall that under the assumptions on equal covariance, equal costs, and equal priors, the sample classification rule for multi-classes that minimizes ECM is
where
In Fisher LDA, assume \(r = p\), let the eigenvector matrix be \(\boldsymbol{A}_{p \times p} = [\boldsymbol{a} _1, \boldsymbol{a} _2, \ldots, \boldsymbol{a} _p]\), then we have
In the Fisher LDA classification rule, the distance can be written as
Therefore, the Fisher LDA classification rule is
which is exactly the linear classification rule for Gaussian Data, under those assumptions.